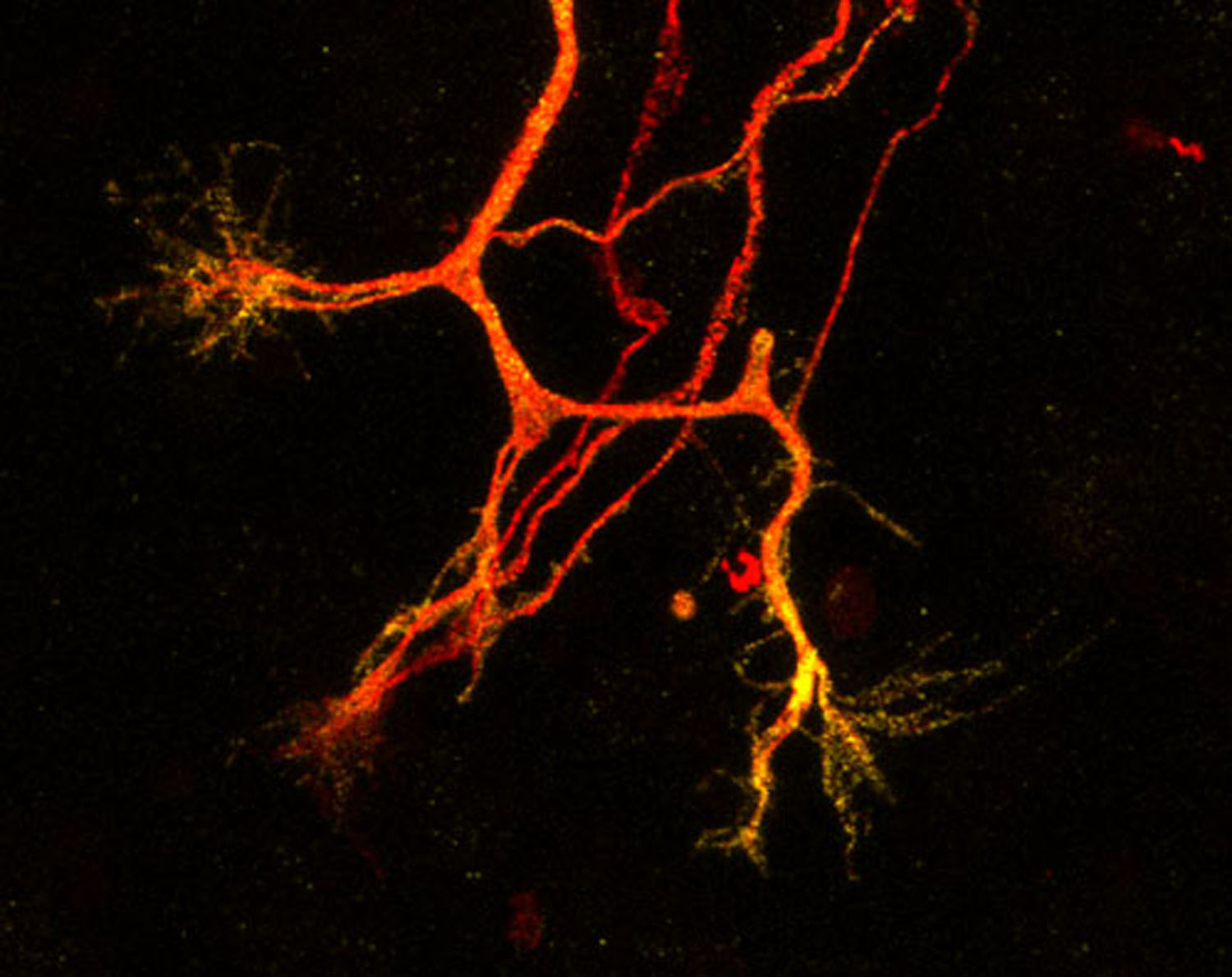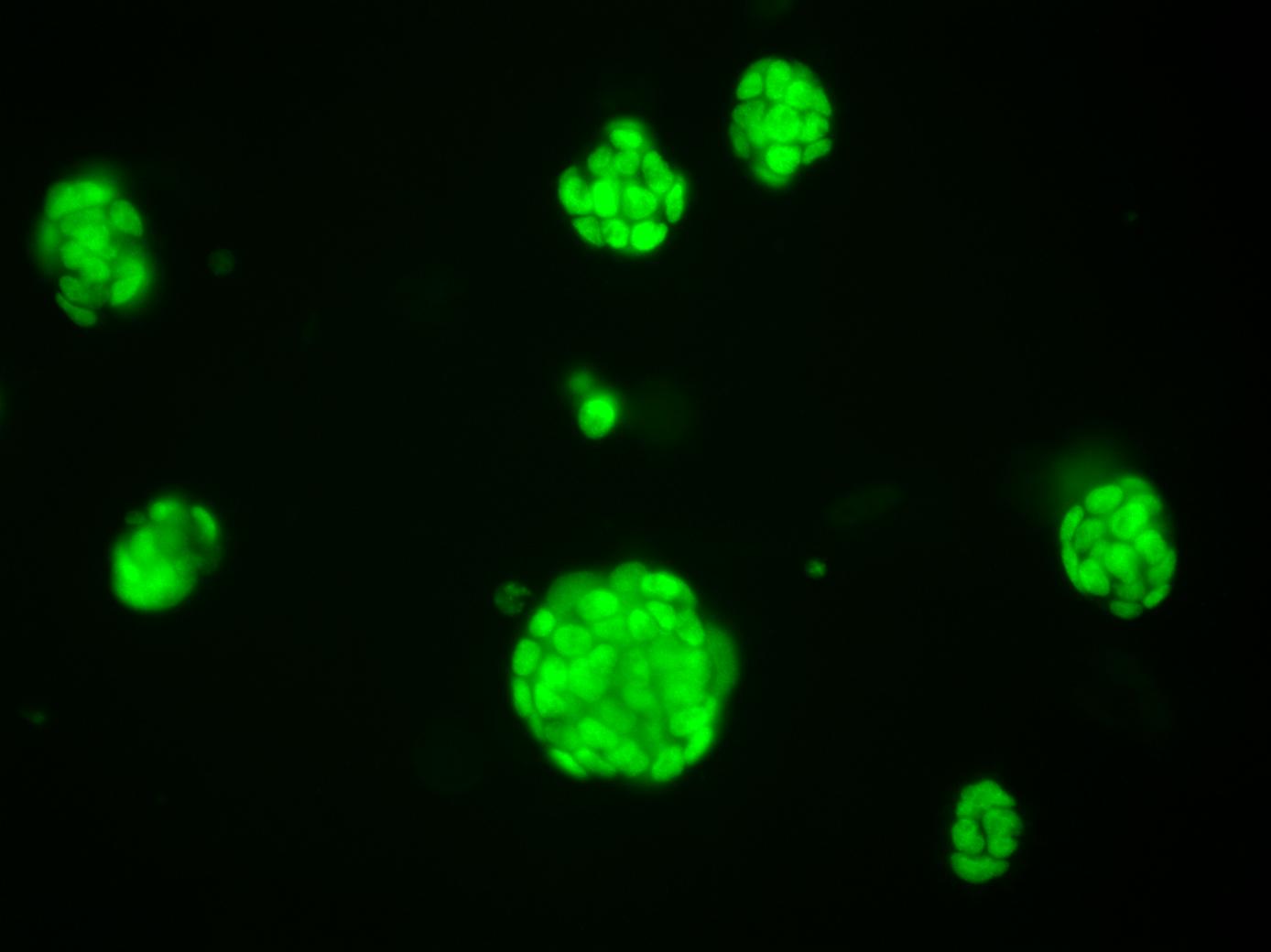
The Laman Lab's mission is to understand and devise novel treatments for diseases caused by deregulation of ubiquitin ligases.
Research in our lab focuses on the study of ubiquitin ligases. They are remarkably multi-functional proteins and touch on aspects of biology ranging from development and regulation of the cell cycle to the immune system and cell signalling. Given such central roles, their malfunctioning is linked with many pathologies, such as cancer, neurodegenerative, and infectious diseases. Our research has branched into many diverse areas, including developing novel therapeutics against ubiquitin ligases, but our focus on these enigmatic enzymes is the common link for our group.
Our laboratory is based on the Downing Site in Tennis Court Road in central Cambridge with access to the nearby Institutes and Departments in the School of Biological Sciences, the city centre and the Colleges.
Phone
+44 (0)1223 333722
hl316@cam.ac.uk
Locations
Department of Pathology,
Division of Cellular and Molecular Pathology,
University of Cambridge Tennis Court Road
Cambridge CB2 1QP
Link to Dept of Pathology
Link to Heike Laman (Pathology)
Clare College,
Trinity Lane,
Cambridge CB2 1TL
Link to Clare College
Research
Ubiquitin ligases are enzymes that decorate proteins in the cell with ubiquitin, a relatively large (76 amino acid) peptide. This modification with ubiquitin on a protein can signal many different things to the target protein, A well-described effect brought about by ubiquitination is that it changes a protein's stability, but other effects include changes to protein activity or localisation, movement of organelles or waste material within cells, and changes to protein interactions with binding partners. My laboratory is particularly intrigued by these less well-studied ubiquitin-mediated signals. We are also interested in ubiquitin signalling within different contexts, as we have discovered ubiquitin networks vary from cell to cell and under stress conditions. We use cell lines, 3D organoids and mouse models to study the molecular basis for this. We want to understand the dynamic nature and complexity of ubiquitin signalling, and invent novel therapeutics exploiting ubiquitin ligase enzymology and their critically regulated pathways.
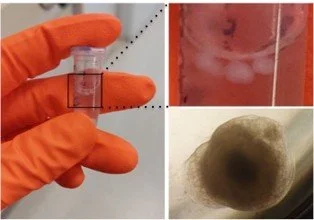
Growing midbrain organoids in the lab.
The fascinating puzzle of the biology of Fbxo7/PARK15: from male sterility to cancer to Parkinson's disease.
Fbxo7, is one of ~70 substrate-recruiting adaptors for the SCF family of E3 ubiquitin ligases. In 1997, we discovered it has the unusual function of acting as a scaffold, promoting the binding of cyclin-dependent kinase 6 (CDK6) with its activators, the D-type cyclins. Their activity is important in driving certain types of cancers, including blood cancers. We use cell lines and 3D organoid cultures to understand the tissue-specific pathologies, and for a more physiologically-relevant studies, we have created multiple mouse models that are deficient in the expression of Fbxo7 either in the whole mouse or in specific cell types. With these tools and systems, we investigate these varied pathologies, including male sterility, anaemia and thymic hypoplasia, stemming from the Fbxo7 deficiency.
A surprising aspect of Fbxo7 biology emerged in 2008 when mutations in Fbxo7 were discovered to be associated with early onset, atypical forms of parkinsonism. We also investigate the loss of its expression in dopaminergic neurons, the cells that die in this disease. We are investigating the molecular basis for the different pathologies caused by the loss of Fbxo7 in these highly divergent cell types. This tissue specificity is not just a biological curiosity - understanding why Fbxo7 loss kills neurons and has roles in cancer is essential for developing safe, targeted therapies for different diseases.
Nanobody interactions with target proteins.
Targeting ubiquitin ligases and ubiquitinated proteins with novel biotherapeutic approaches.
The discovery of camelid-derived antibodies known as nanobodies offers researchers all the specificity and affinities of conventional antibodies in a compact, highly-stable, single domain. Nanobodies are incredibly powerful research tools, as they can be expressed inside of cells, and therefore recognize their antigens in live cells. So they can be engineered, for example, as fusion proteins to fluorescent proteins to be used to visualize proteins in cells. We use nanobodies to define and test the functional significance of interactions between ubiquitin ligases and their substrates. We are investigating using nanobodies as the ligands in biological PROTACs (PRoteolysis TArgeting Chimeras), and we collaborate with AstraZeneca in the design of some of our PROTACs.
In a complementary approach, we are investigating the possibility of specifically interfering with the capacity of ubiquitin ligases to modify their substrates. We collaborate with Laura Itzhaki and David Spring who, based on structural information, produce stapled peptides that mimic the conformation of docked substrates in ubiquitin ligases. We test their biochemical and cellular effects on the ubiquitin ligases.
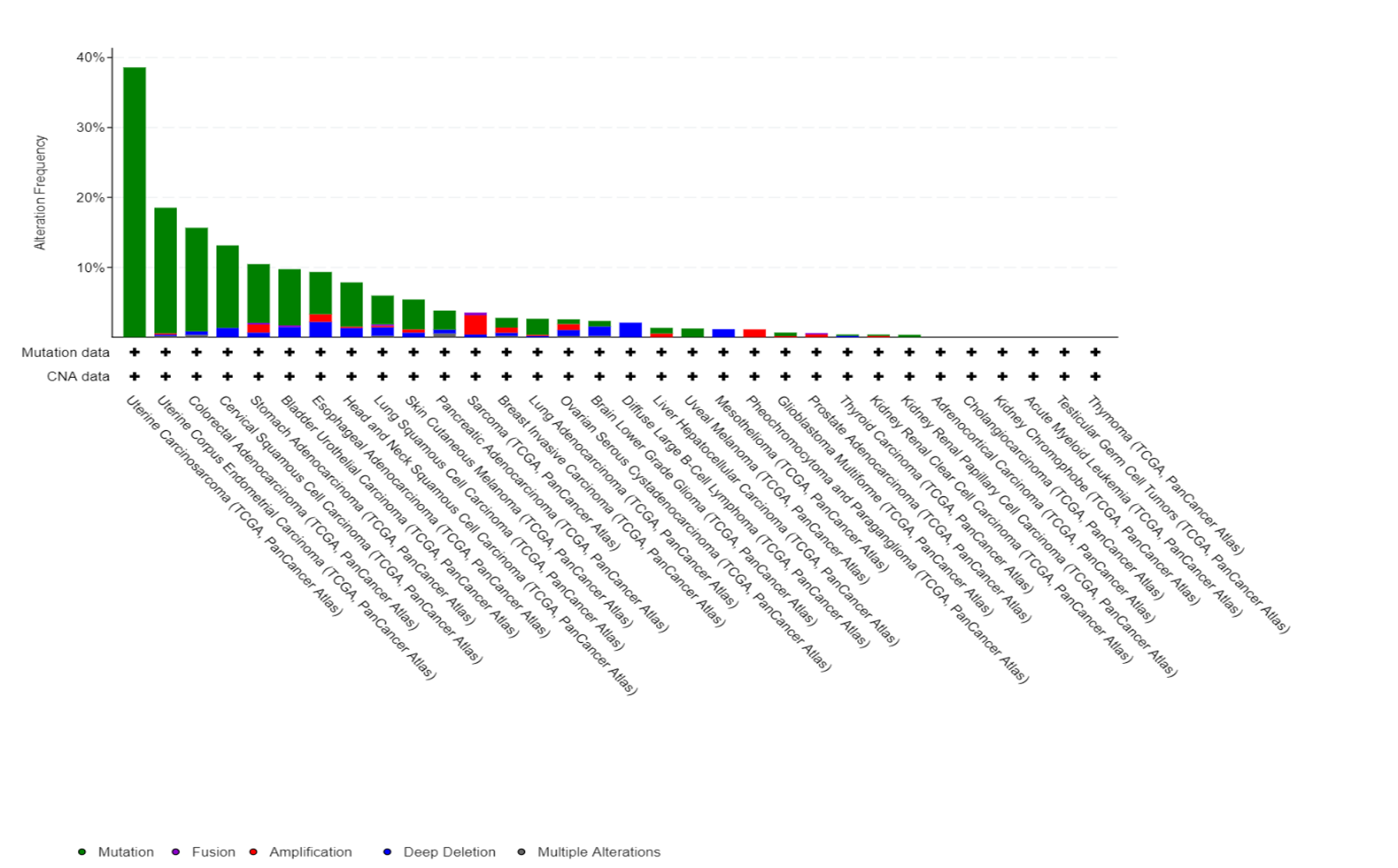
Fbxw7 alterations in TCGA database
Deregulated SCF networks in epithelial cancers.
Fbxw7 is another substrate-recruiting adaptors for the SCF family of E3 ubiquitin ligases and is a well-known tumour suppressor that promotes the degradation of proteins, like cyclin E and Myc. Cyclin E is one of the key proteins that drives cells into division, and its overexpression causes replication stress and genomic instability in many cancers. While cyclin E is normally degraded by the SCF-Fbxw7 ubiquitin ligase, we discovered that many tumors accumulate high cyclin E1 protein levels without gene amplification – suggesting defects in its degradation pathway. Using a multi-omics approach across six cancer types (head and neck, bladder, lung, breast, ovarian, and small cell lung cancer), we are investigating the mechanisms causing cyclin E1 accumulation and identifying patient populations who could benefit from replication stress-targeting therapies. In collaboration with AstraZeneca, we are exploring how cyclin E1 overexpression predicts responsiveness to checkpoint inhibitors and other targeted therapies. Understanding why certain cancers cannot properly degrade cyclin E1 could reveal new therapeutic vulnerabilities and improve patient selection for emerging cancer treatments
Publications, Datasets, & Reagents
Restoration of Fbxo7 expression in dopaminergic neurons restores tyrosine hydroxylase in a mouse model of Parkinson’s disease. Al Rawi S, Tyers P, Barker RA, Laman H. biorxiv 2024 Oct. Link.
Study of an FBXO7 patient mutation reveals Fbxo7 and PI31 co-regulate proteasomes and mitochondria. Al Rawi S*, Simpson L*, Agnarsdóttir G, McDonald NQ, Chernuha V, Elpeleg O, Zeviani M, Barker RA, Spiegel R^, Laman H.^ FEBS J 2024 Feb. Link.
Development of constrained peptide inhibitors targeting an oncogenic E3 ubiquitin ligase. Zenkevičiūtė G, Xu W, Iegre J, Seki H, Tan YS, Rowling PJE, Ferrer F, Verma C, Spring DR, Laman H, Itzhaki LS. biorxiv 2023 May. Link
E3 Ubiquitin Ligases: From Structure to Physiology to Therapeutics, Volume II. Licchesi J, Laman H, Ikeda F, Ferguson F. Bolanos-Garcia V. Front Physiol. 2022. doi.org/10.3389/fphys.2022.1038793 . eCollection. Link.
Fbxo7 promotes Cdk6 activity to inhibit PFKP and glycolysis in T cells. Harris R, Yang M, Schmidt C, Royet C, Singh S, Natarajan A, Morris M, Frezza C, Laman H. J Cell Biol. 2022 Jun 7. doi: 10.1083/jcb.202203095. Link. Spotlight on our article. Link.
A FBXO7/EYA2-SCFFBXW7 axis promotes AXL-mediated maintenance of mesenchymal and immune evasion phenotypes of cancer cells. Shen JZ, Qiu Z, Wu Q, ZhangG, Harris R, Sun D, Rantala J, Barshop WD, Zhao L, Lv D, Won KA, Wohlschlegel J, Sangfelt O, Laman H, Rich JN, Spruck C. Mol Cell. Mar 17;82(6):1123-1139.e8. doi: 10.1016/j.molcel.2022.01.022. Epub 2022 Feb 18. Link.
Laman Lab Plamids available on Addgene! Link.
Analysis of the FBXO7 promoter reveals overlapping Pax5 and c-Myb binding sites functioning in B cells. Harris R, Randle SJ, Laman H. Biochem Biophys Res Comm 2021 Mar 11.doi: 10.1016/j.bbrc.2021.03.052. Epub 2021 Mar 25. Link.
The FBXL family of F-box proteins: variations on a theme. Mason BJ & Laman H. Open Biol. 2020 Nov;10(11):200319. doi: 10.1098/rsob.200319. Epub 2020 Nov 25. Link.
E3 Ubiquitin Ligases: From Structure to Physiology. Licchesi JDF, Laman H, Ikeda F, Bolanos-Garcia VM. Front Physiol. 2020. Nov 26;11:621053. doi: 10.3389/fphys.2020.621053. eCollection.. Link.
The E3 ubiquitin ligase SCF(Fbxo7) mediates proteasomal degradation of UXT isoform 2 (UXT-V2) to inhibit the NF-kappa B signaling pathway. Spagnol V, Oliveira CAB, Randle SJ, Passos PMS, Correia CRSTB, Simaroli NB, Oliveira JS, Mevissen TET, Medeiros AC, Gomes MD, Komander D, Laman H, Teixeira FR. Biochim Biophys Acta. 2020 Sep 30;1865(1):129754. doi: 10.1016/j.bbagen.2020.129754. Link.
Nedd8 hydrolysis by UCH proteases in Plasmodium parasites. Karpiyevich M, Adjalley S, Mol M, Ascher DB, Mason B, van der Heden van Noort GJ, Laman H, Ovaa H, Lee MCS, Artavanis-Tsakonas K. PLoS Pathog. 2019 Oct 28;15(10):e1008086. doi: 10.1371/journal.ppat.1008086. Link. PDF.
A conserved requirement for Fbxo7 during male germ cell cytoplasmic remodelling. Rathje CC, Randle SJ, Al Rawi S, Skinner BM, Rajamundar A, Nelson DE, Johnson EEP, Bacon J, Vlazak M, Affara NA, Ellis PJ, Laman H. Front Physiol. 2019. Oct 10;10:1278. doi: 10.3389/fphys.2019.01278. Link. PDF.
Fbxl17 is rearranged in breast cancer and loss of its activity leads to increased global O-GlcNAcylation. Mason B, Flach S, Teixeira, FR, Manzano Garcia R, Rueda OM, Abraham JE, Caldas C, Edwards PAW. Laman H. Cell Mol Life Sci. 2019 Sept 27. Link. PDF.
Loss of FBXO7 results in a Parkinson’s-like dopaminergic degeneration via an RPL23-MDM2-TP53 pathway. Stott SRW*, Randle SJ*, al Rawi S, Rowicka PA, Harris R, Mason B, X.ia J, Dalley, JW, Barker RA, Laman H. J Pathol. 2019 Oct;249(2):241-254. Link. PDF.
Opposing effects on the cell cycle of T lymphocytes by Fbxo7 via Cdk6 and p27. Patel SP, Randle SJ, Gibbs S, Cooke A, Laman H. Cell Mol Life Sci. 2017 Apr;74(8):1553-1566 . Link. PDF
Structure and function of Fbxo7/PARK15 in Parkinson's disease. Randle SJ, Laman H. Curr Protein Pept Sci. 2017;18(7):715-724. Link. PDF.
Gsk3β and Tomm20 are substrates of the SCF-Fbxo7/PARK15 ubiquitin ligase associated with Parkinson's disease. Teixeira FR^, Randle SJ^, Patel SP^, Mevissen TE, Zenkeviciute G, Koide T, Komander D, Laman H. Biochem J. 2016 Oct 15;473(20):3563-3580. ^First author. Link. PDF. Supp.
F-box protein interactions with the hallmark pathways in cancer. Randle SJ, Laman H. Semin Cancer Biol. 2016 Feb;36:3-17. Link. PDF.
Defective erythropoiesis in a mouse model of reduced Fbxo7 expression due to decreased p27 expression. Randle SJ, Nelson DE, Patel SP, Laman H. J Pathol. 2015 Jun;237:263-272. Link. PDF. Supp.
Beyond ubiquitination: the atypical functions of Fbxo7 and other F-box proteins. Nelson DE, Randle SJ, Laman H. Open Biol. 2013 Oct 9;3(10):130131. Link. PDF.
The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Burchell VS^, Nelson DE^, Sanchez-Martinez A^, Delgado-Camprubi M, Ivatt RM, Pogson JH, Randle SJ, Wray S, Lewis PA, Houlden H, Abramov AY, Hardy J, Wood NW, Whitworth AJ*, Laman H*, Plun-Favreau H*. Nat Neurosci. 2013 Sep;16(9):1257-65. ^First author; *Senior corresponding author. Link. PDF.
Identification of F-box only protein 7 as a negative regulator of NF-kappaB signalling. Kuiken HJ, Egan DA, Laman H, Bernards R, Beijersbergen RL, Dirac AM. J Cell Mol Med. 2012 Sep;16(9):2140-9. Link. PDF.
Exploring the interaction between siRNA and the SMoC biomolecule transporters: implications for small molecule-mediated delivery of siRNA. Gooding M, Tudzarova S, Worthington RJ, Kingsbury SR, Rebstock AS, Dube H, Simone MI, Visintin C, Lagos D, Quesada JM, Laman H, Boshoff C, Williams GH, Stoeber K, Selwood DL. Chem Biol Drug Des. 2012 Jan;79(1):9-21. Link. PDF.
Expression of Fbxo7 in haematopoietic progenitor cells cooperates with p53 loss to promote lymphomagenesis. Lomonosov M, Meziane el K, Ye H, Nelson DE, Randle SJ, Laman H. PLoS One. 2011;6(6):e21165. Link. PDF.
Knockdown of Fbxo7 reveals its regulatory role in proliferation and differentiation of haematopoietic precursor cells. Meziane el K^, Randle SJ^, Nelson DE, Lomonosov M, Laman H. J Cell Sci. 2011 Jul 1;124(Pt 13):2175-86. ^First author. Link. PDF.
A Competitive binding mechanism between Skp1 and exportin 1 (CRM1) controls the localization of a subset of F-box proteins. Nelson DE, Laman H. J Biol Chem. 2011 Jun 3;286(22):19804-15. Link. PDF.
Structure of a conserved dimerization domain within the F-box protein Fbxo7 and the PI31 proteasome inhibitor. Kirk R, Laman H, Knowles PP, Murray-Rust J, Lomonosov M, Meziane el K, McDonald NQ. J Biol Chem. 2008 Aug 8;283(32):22325-35. Link. PDF
Small-molecule mimics of an alpha-helix for efficient transport of proteins into cells.Okuyama M^, Laman H^, Kingsbury SR^, Visintin C, Leo E, Eward KL, Stoeber K, Boshoff C, Williams GH, Selwood DL. ^First author. Nat Methods. 2007 Feb;4(2):153-9. Link. PDF
Fbxo7 gets proactive with cyclin D/cdk6. Laman H. Cell Cycle. 2006 Feb;5(3):279-82. Link. PDF.
Transforming activity of Fbxo7 is mediated specifically through regulation of cyclin D/cdk6. Laman H, Funes JM, Ye H, Henderson S, Galinanes-Garcia L, Hara E, Knowles P, McDonald N, Boshoff C. EMBO J. 2005 Sep 7;24(17):3104-16. Link. PDF.
Regulation of growth signalling and cell cycle by Kaposi's sarcoma-associated herpesvirus genes. Direkze S, Laman H. 2004 Dec;85(6):305-19. Link. PDF.
RNA interference: a potential tool against Kaposi's sarcoma-associated herpesvirus. Godfrey A, Laman H, Boshoff C. Curr Opin Infect Dis. 2003 Dec;16(6):593-600. Link.
Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Coverley D, Laman H, Laskey RA. Nat Cell Biol. 2002 Jul;4(7):523-8. Link. PDF
Cyclin-mediated export of human Orc1. Laman H, Peters G, Jones N. Exp Cell Res. 2001 Dec 10;271(2):230-7. Link. PDF
Is KSHV lytic growth induced by a methylation-sensitive switch? Laman H, Boshoff C. Trends Microbiol. 2001 Oct;9(10):464-6. Link. PDF.
Viral cyclin-cyclin-dependent kinase 6 complexes initiate nuclear DNA replication. Laman H, Coverley D, Krude T, Laskey R, Jones N. Mol Cell Biol. 2001 Jan;21(2):624-35. Link. PDF.
Crystal structure of a gamma-herpesvirus cyclin-cdk complex. Card GL, Knowles P, Laman H, Jones N, McDonald NQ. EMBO J. 2000 Jun 15;19(12):2877-88. Link. PDF.
Viral-encoded cyclins. Laman H, Mann DJ, Jones NC. Curr Opin Genet Dev. 2000 Feb;10(1):70-4. Link. PDF.
Modulation of p27(Kip1) levels by the cyclin encoded by Kaposi's sarcoma-associated herpesvirus.Mann DJ, Child ES, Swanton C, Laman H, Jones N. EMBO J. 1999 Feb 1;18(3):654-63. Link. PDF.
Disturbance of normal cell cycle progression enhances the establishment of transcriptional silencing in Saccharomyces cerevisiae. Laman H, Balderes D, Shore D. Mol Cell Biol. 1995 Jul;15(7):3608-17. Link. PDF.
Identification of a nitrogen-regulated promoter controlling expression of Klebsiella pneumoniae urease genes. Collins CM, Gutman DM, Laman H. Mol Microbiol. 1993 Apr;8(1):187-98. Link.
PI Biography
Professor Heike Laman
Heike Laman is Head of the Department of Pathology and Professor in Cellular and Molecular Biology at the University of Cambridge, where she leads research on ubiquitin ligases and their diverse cellular signals. https://www.path.cam.ac.uk/.
Her research career began at the University of Miami, where she graduated cum laude and with General Honours in 1990 with a BSc in Microbiology & Immunology and a double minor in Chemistry and Biology. Her studies were distinguished by elections to the academic honour societies, Phi Beta Kappa and Phi Beta Phi. As an undergraduate research assistant with Dr Carleen Collins, she sequenced the entire urease operon in Klebsiella pneumoniae and identified its regulatory elements. This was a tour-de-force of sequencing and analysis in the pre-genomics era. This early success in the lab led her to pursue graduate studies at Columbia University.
At Columbia, she worked with Dr David Shore investigating heterochromatin assembly in yeast, earning her MA, MPhil and PhD degrees. Having been awarded a prestigious Fellowship from the Imperial Cancer Research Fund (now subsumed into the Francis Crick Institute), she then moved to the UK for postdoctoral positions, where she studied viral cyclins with Prof Nic Jones and cell cycle regulation with Dr Gordon Peters. This work revealed how viruses manipulate cellular control mechanisms – insights that influenced her subsequent research. She continued as a Senior Research Fellow at UCL's Wolfson Institute with Prof Chris Boshoff, supported by funding from the Association for International Cancer Research.
In 2005, Professor Laman was awarded a Research Fellowship to establish her independent research group investigating ubiquitin ligases in the Department of Pathology at the University of Cambridge. Her laboratory has uncovered how these enzymes create diverse cellular signals beyond protein degradation, affecting protein activity, localization, and interactions. This work spans basic mechanisms to disease applications, including collaborations with industrial partners.
She was elected to the Fellowship of Clare College in 2014 and is Director of Studies (Part IB Biology of Disease; Part II Pathology; Part II Genetics), and is the Graduate Admissions Tutor, and a Trustee of the College.
Our Team
We are always seeking talented, enthusiastic, and curious minds to join our group. Post-doctoral candidates are encouraged to get in touch, sending your CV, to enquire about opportunities to join our lab.
Current Members
Finn Snow
Finn joined the lab in October 2024 to begin his MRC-ITTP-funded PhD studentship in collaboration with AstraZeneca in targeted protein degradation and ubiquitination. He graduated from the University of Bath with a first class BSc. degree in Biochemistry, spending a placement year at GSK working on protein target generation. Since developing a passion for pharmaceutical research, he completed a MSc. Drug Design degree (distinction) from University College London, where his project focused on developing a small molecule diagnostic for alpha-1 antitrypsin deficiency. His PhD research will mainly focus on understanding the unique off-target mechanisms of Proteolysis Targeting Chimeras (PROTACs).
Sara al Rawi
Born in Paris, Sara joined the lab in 2018 to start a Parkinson's UK-funded research project investigating the neuroprotective roles of Fbxo7 with a focus on an early stages of Parkinson's disease. Previously, Sara obtained a Master degree in cellular and molecular biology with a specialization in proteomics from the Pierre and Marie Curie University, in Paris. After being awarded her degree, Sara began working as a research assistant to investigate how sperm derived mitochondria are degraded in the early embryo using C. elegans as a model. This work led to the discovery of the autophagy pathway in this specific degradation event. Sara was awarded an AXA-Academie des Sciences award from the French Science academy for her work. Her PhD research was under the supervision of Dr Vincent Galy at the Pierre and Marie Curie University and focussed on discovering how sperm-derived mitochondria are specifically recognized and targeted by the autophagy machinery.
Sophie Willis
Sophie joined the lab in October 2020 to start a PhD investigating the role of ubiquitination in different subtypes of Cyclin E high cancers. She graduated from the University of Leeds in 2016 with a Bachelor of Science (Hons) in Genetics (Industrial), spending her placement year at MedImmune investigating immune cell populations in pancreatic cancer using immunohistochemistry and NanoString gene expression assays. Since graduating, she has worked at AstraZeneca, developing immunohistochemistry and RNA in situ hybridisation assays to support pharmacodynamic assessments and patient selection strategies on a range of clinical projects.
Hanna Bjone
Hanna joined the lab in October 2021 to start her PhD funded by Aker Scholarship (Norway) to investigate the neuroprotective effects of Fbxo7 in Parkinson's disease focusing on its effects on Cdk6 activity and intracellular trafficking. Born in Oslo, Hanna moved to the UK for her undergraduate studies and graduated from Durham University with a first class integrated Master’s degree in biological sciences. Her final year project focused on the production and optimization of recombinant insecticidal fusion proteins. Prior to coming to Cambridge, she spent a year at the University of Oslo completing a course in computer science in bioinformatics.
Guðrún Diljá Agnarsdóttir
Guðrún started in the lab in October 2022 for her MPhil and is continuing now to study for her PhD. She is investigating the ubiquinome of the pathogenic missense mutation L250P in Fbxo7, which is associated with atypical parkinsonism. She graduated in 2022 from the University of Iceland with a first class BSc degree in Biochemistry and Molecular Biology. During the final year of her degree, she worked as a research associate in the Biological Materials Facility at deCODE Genetics in Iceland. Guðrún also completed her undergraduate dissertation project at deCODE, using their vast amount of omics data to examine the phenotypic impact of rare genetic variants in the human CYP2R1 gene.
Libby Wiseman
Libby joined the lab in October 2023 to begin her PhD, investigating the utility of nanobodies and targeted protein degradation in novel biological therapies for Myc driven cancers, such as neuroblastoma. She graduated in 2018 from Oxford Brookes University with a first class Bachelor of Science (Hons) degree in Biomedical Science, where her final year project focused on cellular senescence following radiotherapy. In 2021, Libby became a Scientist at AstraZeneca, where she works on validation of new cancer biomarkers using translational techniques such as immunohistochemistry, with the aim to achieve better understanding of patient response to treatment and mechanisms of resistance.
Min Zhang
Min joined the lab in February 2024 to start her postdoctoral research on how Fbxo7 and PI31 control sperm morphogenesis and male fertility. Previously, Min obtained a Master degree in Clinical Veterinary Medicine focusing on nutritional and metabolic diseases in perinatal dairy cows at Jilin University, China. Min moved to The Netherlands in October 2016 and started her PhD in Biochemistry and Reproductive Biology at Utrecht University. Her PhD research characterized the involvement of CRISP2 in the organization of dense protein structures in porcine sperm.
Daniel Tikhomirov
Daniel joined the lab in April 2025 to continue the third year of his PhD, focusing on the off-target toxicity of the antipsychotic aripiprazole in both 2D and 3D hiPSC-derived midbrain organoids. He is also working on a collaborative project with bit.bio, investigating how mitochondrial heteroplasmy may influence a cell’s ability to reprogram and differentiate. Prior to his PhD at the University of Cambridge, Daniel graduated with First Class Honours in Medical Sciences from the University of Edinburgh. During his studies, he worked as a research assistant in Prof. Tilo Kunath’s lab, where he contributed to the development of a non-invasive method for monitoring the differentiation of midbrain dopaminergic neurons.
Ting Deng
Ting joined the lab in April 2025 as a second-year PhD student. Her research investigates the role of Fbxo7 in regulating the interferon-gamma (IFN-γ)-induced integrated stress response (ISR), with a particular focus on dopaminergic neurons. Prior to her PhD, she earned her Master’s degree from Tsinghua University, where she worked on the development of peptide-based therapeutics and immune cell therapies for colorectal cancer. Her broader research interests include mitochondrial homeostasis, translational control, and the molecular mechanisms underlying neurodegeneration
Aristos Song
Aristos joined the lab in September 2025 to pursue her MPhil, investigating how the Parkinson's disease-associated protein FBXO7 regulates intracellular trafficking through ubiquitination of intracellular trafficking proteins. She graduated from UCL with first-class honors in Biomedical Science. During her final year, she worked at the Francis Crick Institute on an AstraZeneca-funded project developing novel tools to assess proteasome-dependent and autophagy-dependent degradation of undruggable targets.
Past Lab Members
Dr Rebecca Harris is a Senior Scientist at Amphista Therapeutics.
Dr Lorna Simpson has joined Muscular Dystrophy UK.
Dr Andrei Smid is a Senior Scientist at Revvity.
Dr Paulina Rowicka is a post-doctoral fellow at Harvard Medical School.
Dr Bethany (nee Mason) South is a Senior Scientist at Artios Pharma Ltd.
Dr Lisa Kent is an Independent Consultant at StaKe Consulting Ltd
Dr Suzanne Randle is a Associate Director in Oncology Safety Science at AstraZeneca in Cambridge
Dr Shachi Patel is Associate Principal at ClearView Healthcare Partners
Dr Felipe Teixeira is Principal Investigator in Cellular Biochemistry at the Department of Genetics and Evolution at the Federal University of Sao Carlos in Brazil.
Dr David Nelson is an Associate Professor at Middle Tennesee State University in the USA.
Dr El Kahina Meziane is Head of Biology at the International School in London, UK.
Dr Mikhail Lomonsov is Head of Research at Ivix Ltd.
Collaborators
Prof. Roger Barker: Department of Clinical Neurosciences, University of Cambridge, Link.
Dr. Erwin de Genst: AstraZeneca, Discovery Science, AstraZeneca. Link.
Dr. Laura Itzhaki: Department of Pharmacology, University of Cambridge. Link.
Prof. Kathryn Lilley: Department of Biochemistry, University of Cambridge. Link.
Dr. David Spring: Department of Chemistry, University of Cambridge. Link.
Prof Felipe Teixeira: Department of Genetics and Evolution, Universidad Federal Sao Carlos. Link.
Prof Massimo Zeviani: Veneto Institute for Molecular Medicine (VIMM), Padua. Link.
Links
Some of our favourite review journals
Current Opinion in Genetics and Development
Current Opinion in Infectious Disease
Nature Reviews Molecular Cell Biology
Resources
Addgene (Laman Lab plasmids)
American Type Culture Collection (ATCC)
Bioinformatics Tools
Biomednet
BLAST
elm Database
Embryo Visualization Page
Ensembl Genome Browser
Entrez
Human Embryonic Development
Jackson Laboratory
Journal Citation Reports
MethPrimer
The Mouse Atlas Project
Primer-Blast
Protein Data Bank
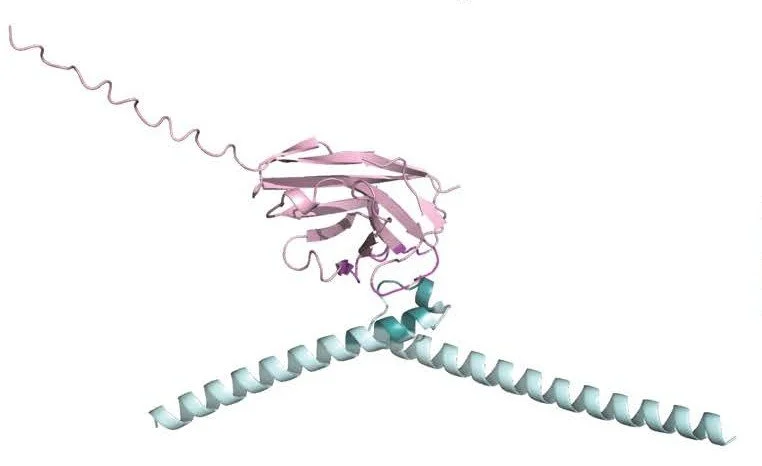
The dimer conformation arising from beta-sheet interactios of the FP domain of PI31.
Contact Us
Laman Lab
University of Cambridge
Department of Pathology
Tennis Court Road
Cambridge CB2 1QP
United Kingdom