Research
Ubiquitin ligases are enzymes that decorate proteins in the cell with ubiquitin, a relatively large (76 amino acid) peptide. This modification with ubiquitin on a protein can signal many different things to the target protein, A well-described effect brought about by ubiquitination is that it changes a protein's stability, but other effects include changes to protein activity or localisation, movement of organelles or waste material within cells, and changes to protein interactions with binding partners. My laboratory is particularly intrigued by these less well-studied ubiquitin-mediated signals. We are also interested in ubiquitin signalling within different contexts, as we have discovered ubiquitin networks vary from cell to cell and under stress conditions. We use cell lines, 3D organoids and mouse models to study the molecular basis for this. We want to understand the dynamic nature and complexity of ubiquitin signalling, and invent novel therapeutics exploiting ubiquitin ligase enzymology and their critically regulated pathways.
Growing midbrain organoids in the lab.
The fascinating puzzle of the biology of Fbxo7/PARK15: from male sterility to cancer to Parkinson's disease.
Fbxo7, is one of ~70 substrate-recruiting adaptors for the SCF family of E3 ubiquitin ligases. In 1997, we discovered it has the unusual function of acting as a scaffold, promoting the binding of cyclin-dependent kinase 6 (CDK6) with its activators, the D-type cyclins. Their activity is important in driving certain types of cancers, including blood cancers. We use cell lines and 3D organoid cultures to understand the tissue-specific pathologies, and for a more physiologically-relevant studies, we have created multiple mouse models that are deficient in the expression of Fbxo7 either in the whole mouse or in specific cell types. With these tools and systems, we investigate these varied pathologies, including male sterility, anaemia and thymic hypoplasia, stemming from the Fbxo7 deficiency.
A surprising aspect of Fbxo7 biology emerged in 2008 when mutations in Fbxo7 were discovered to be associated with early onset, atypical forms of parkinsonism. We also investigate the loss of its expression in dopaminergic neurons, the cells that die in this disease. We are investigating the molecular basis for the different pathologies caused by the loss of Fbxo7 in these highly divergent cell types. This tissue specificity is not just a biological curiosity - understanding why Fbxo7 loss kills neurons and has roles in cancer is essential for developing safe, targeted therapies for different diseases.
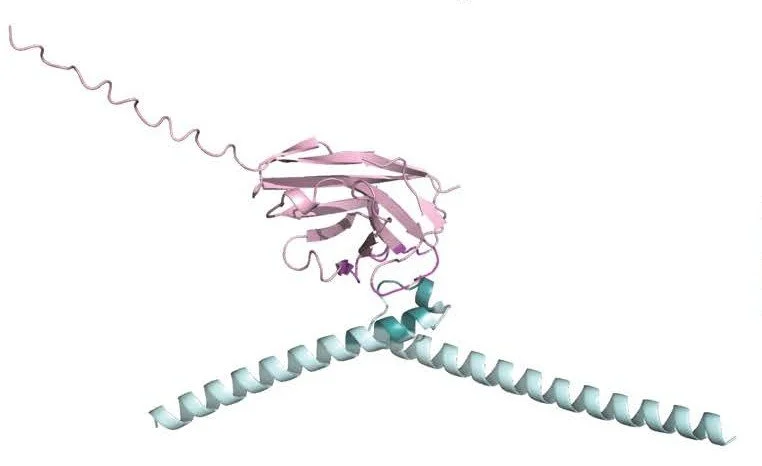
Nanobody interactions with target proteins.
Targeting ubiquitin ligases and ubiquitinated proteins with novel biotherapeutic approaches.
The discovery of camelid-derived antibodies known as nanobodies offers researchers all the specificity and affinities of conventional antibodies in a compact, highly-stable, single domain. Nanobodies are incredibly powerful research tools, as they can be expressed inside of cells, and therefore recognize their antigens in live cells. So they can be engineered, for example, as fusion proteins to fluorescent proteins to be used to visualize proteins in cells. We use nanobodies to define and test the functional significance of interactions between ubiquitin ligases and their substrates. We are investigating using nanobodies as the ligands in biological PROTACs (PRoteolysis TArgeting Chimeras), and we collaborate with AstraZeneca in the design of some of our PROTACs.
In a complementary approach, we are investigating the possibility of specifically interfering with the capacity of ubiquitin ligases to modify their substrates. We collaborate with Laura Itzhaki and David Spring who, based on structural information, produce stapled peptides that mimic the conformation of docked substrates in ubiquitin ligases. We test their biochemical and cellular effects on the ubiquitin ligases.
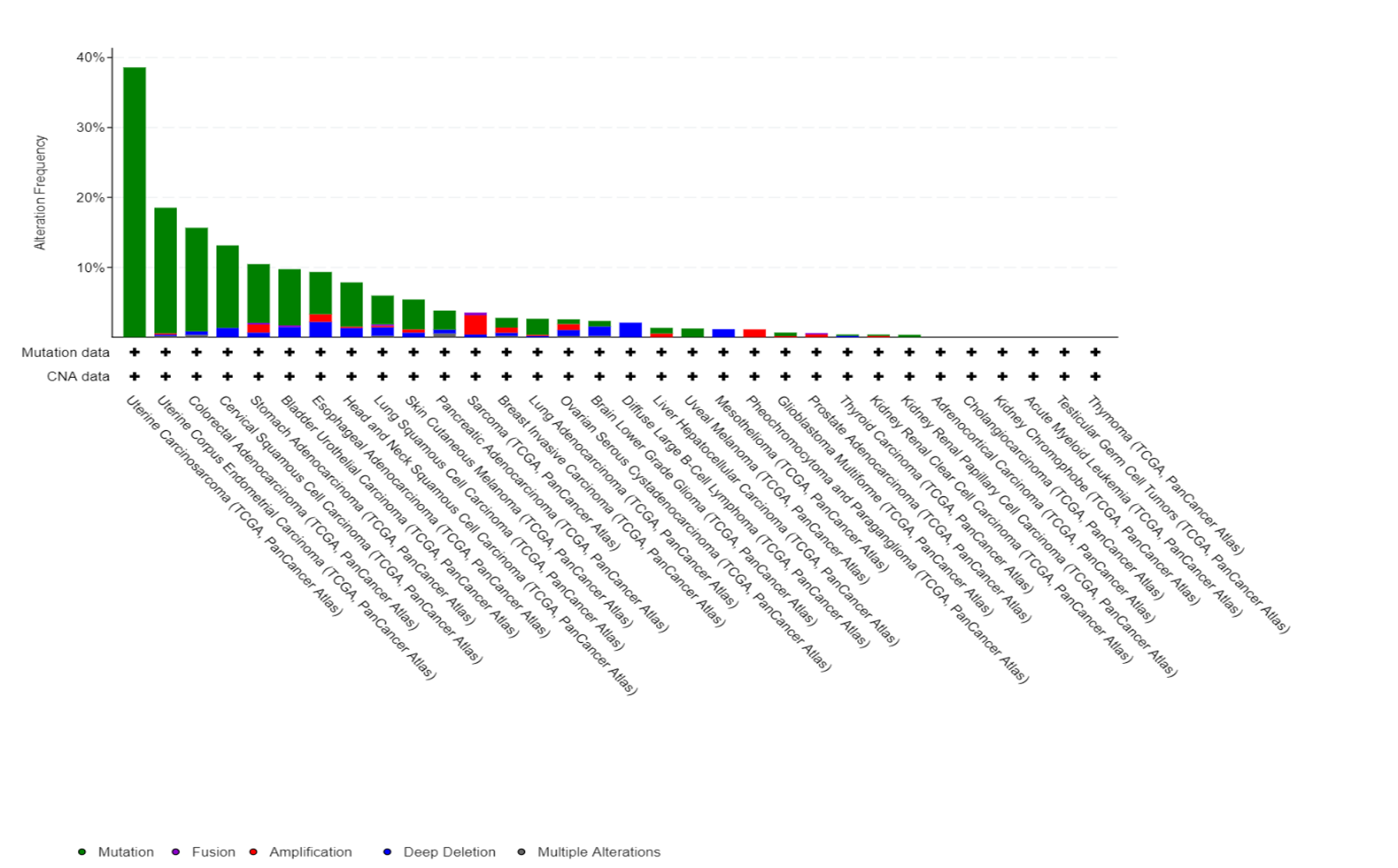
Fbxw7 alterations in TCGA database
Deregulated SCF networks in epithelial cancers.
Fbxw7 is another substrate-recruiting adaptors for the SCF family of E3 ubiquitin ligases and is a well-known tumour suppressor that promotes the degradation of proteins, like cyclin E and Myc. Cyclin E is one of the key proteins that drives cells into division, and its overexpression causes replication stress and genomic instability in many cancers. While cyclin E is normally degraded by the SCF-Fbxw7 ubiquitin ligase, we discovered that many tumors accumulate high cyclin E1 protein levels without gene amplification – suggesting defects in its degradation pathway. Using a multi-omics approach across six cancer types (head and neck, bladder, lung, breast, ovarian, and small cell lung cancer), we are investigating the mechanisms causing cyclin E1 accumulation and identifying patient populations who could benefit from replication stress-targeting therapies. In collaboration with AstraZeneca, we are exploring how cyclin E1 overexpression predicts responsiveness to checkpoint inhibitors and other targeted therapies. Understanding why certain cancers cannot properly degrade cyclin E1 could reveal new therapeutic vulnerabilities and improve patient selection for emerging cancer treatments